top
top
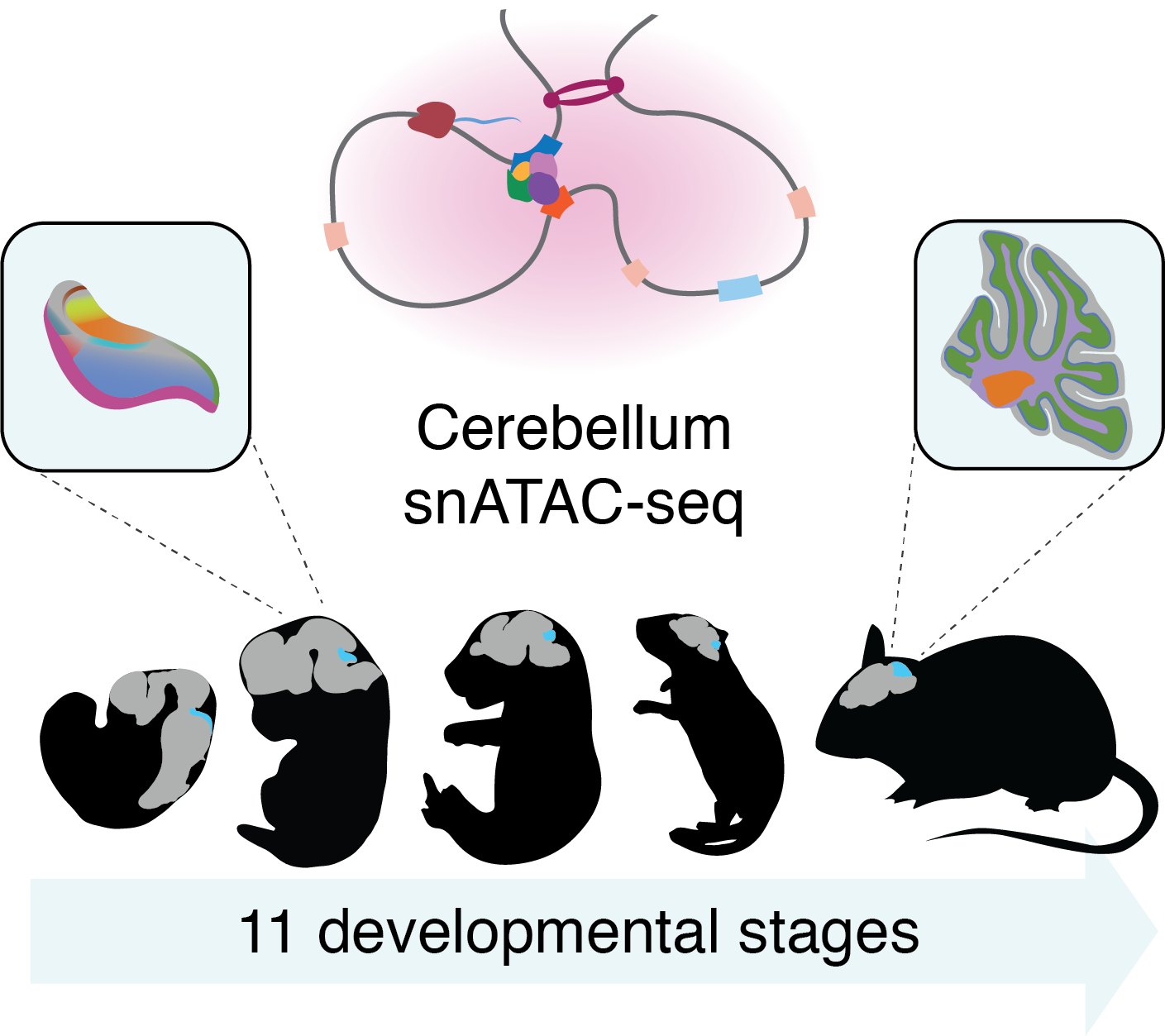
This app allows the exploration of cis-regulatory elements (CREs) active in the developing mouse cerebellum.
If you are using this app, please consult and cite our publication:
Sarropoulos, I., Sepp M. et al. Developmental and evolutionary dynamics of cis-regulatory elements in mouse cerebellar cells. Science (2021)
1. Select a CRE
You can scroll horizontally to explore additional options.
2. Show in genome browser
Your CRE is shown in red. Other robust/non-robust CREs are shown in dark/light blue respectively.
Genomic interactions highlight CRE assignment to target genes in our study (purple/blue: in/out browser view).
Genomic interactions highlight CRE assignment to target genes in our study (purple/blue: in/out browser view).
Genomic region displayed:
Please select a CRE
3. Plot CRE activity
Cell annotation
Not a valid symbol
This option is only available for shared CREs, with robust activity in both species.
Download plot
Please select a CRE
4. Download data
Warning: Some of these files are large, so downloading might take a while.
RDS files contain SummarizedExperiment objects (1.16) that can be read into R using readRDS().
RDS files contain SummarizedExperiment objects (1.16) that can be read into R using readRDS().
Name
Size
Description
(2.0 GB)
CRE by cell matrix with associated information about peaks and cells (mouse).
(1.1 GB)
chromVar motif scores (deviations and Z-scores) by cell matrix (mouse).
(70 MB)
Summary information for mouse CREs (genomic and evolutionary characterization, activity etc).
(50 MB)
Summary information for mouse cells (annotation, sample, clusters etc).
(1 MB)
Summary information for opossum cells (annotation, sample, clusters etc).